3D virus image has the biggest resolution ever
Purdue University team has succeeded in creating virus images with twice the resolution of the past.
Wen Jiang, an assistant professor of biological sciences at Purdue University, led the team to use a single electronic cold microscope technique to get 3D images of viruses with a resolution of 4.5 angstroms (unit of measurement). light wavelength). An average of 1 million angstroms is equivalent to the diameter of a human hair.
Jiang is also a member of Purdue university biology group, saying: 'This is one of the first projects to improve the technique to achieve atomic level resolution. The project has created a breakthrough, allowing us to observe details at a new level in this structure. This is the largest resolution achieved for living organisms of the size of phage. ' He also added that detailed structure of the virus will provide valuable information in the study of disease treatment.
'If we understand how viral parts are gathered together, how they infect host cells, our ability will be enhanced to study treatments. Structural biologists have conducted basic science and provided information to study medical fields. '
Roger Hendrix, a professor of biological sciences at the University of Pittsburgh, said what we learned from the virus could be applied to many other biological systems.
He said: 'Understanding the proteins that make up the virus structure provides us with an inside image of the bio-apparatus that is present throughout our bodies. Achieving a resolution of 4.5 angstrom is indeed a major turning point because this is the first time we can actually observe the polypeptide chain - the 'skeleton' of proteins. We can now see tiny parts as well as mechanisms that help proteins move and interact while performing our complex biological roles. '
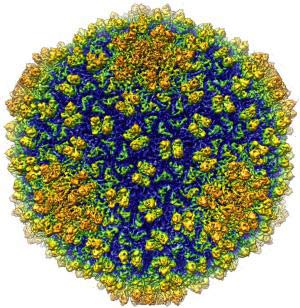
The image of the bacterium Epsilon 15 was studied by assistant professor of biological science Wen Jiang at Purdue.The image of phage may have a resolution of 4.5 angstroms - the largest resolution ever achieved for a living organism the size of phage.(Photo: Wen Jiang Laboratory)
The imaging technique called cryo-EM has the advantage of maintaining the specimen studied in a condition similar to the natural environment . Other techniques commonly used such as X-ray crystallography require manipulation of specimens.
Jiang said: 'This method is very new in molding protein structures in parts of other macromolecules (such as DNA) in nearly original conditions. The specimen is purified in solution almost like the environment in the host cell. The virus is hardened in glass but still alive and able to infect when we study. '
Besides Jiang also has scientist Matthew L. Baker, Joanita Jakana and Wah Chiu of Baylor College of Medicine, scientist Peter R. Weigele and Jonathan King of Massachusetts Institute of Technology. The project is funded by the National Institutes of Health and the National Science Foundation.
Jiang said his team has established a three-dimensional map of the outer protein shell of the phage epsilon 15 - a virus that feeds on bacteria and is also a member of a virus family that is the most common form of life. on the earth.
Other methods of determining the structure cannot be applied to this virus strain. The unsuccessful implementation of crystallization as well as the complexity of the virus strain hindered the evaluation through gene sequencing.
'The new method shows that cryo-EM is completely feasible and this will also be a major step in achieving the full potential of technology. Our goal is to achieve a resolution of 3 to 4 angstroms allowing us to clearly observe the amino acids that make up the protein molecule '.
In electron microscopy, electron beams are used to replace light rays used in conventional microscopic methods. The electrons used in place of light allow much smaller details to be observed through a microscope.
Cryo-EM method cools the sample at a temperature below the freezing level of water. This technique helps minimize electron damage, allowing it to be carried out on specimens for a longer period of time . Once more time is taken with the specimen, we will get more detailed and specific images.
Researchers using cryo-EM have obtained images with a resolution of 6-9 angstrom, but that still does not distinguish the small parts in the structure located 4.5 angstroms apart.
Jiang said: 'To make a protein block in a virus requires a lot of different components. This is like carrying out an operation on a striped blanket. From a distance, stripes line together to make the blanket look like there is only one color. But when you get closer, you can see different stripes. If you use a magnifying glass, you can see the threads that make up the object. The resolution needed to be smaller than the distance between the lines so that two lines can be seen '.
'If you can exaggerate, researchers will see what components are imported as one of the previous photos.'
The Cryo-EM method requires expensive electron microscopy and abundant computer resources. The team used the electron microscope of Baylor College of Medicine. They hope to be able to install the advanced electron microscope at Purdue University in 2009.
In 2006, Purdue University received $ 2 million in support from the National Institutes of Health to buy microscopes. It will be installed at the Hockmeyer Structural Biology department, which is scheduled to open in 2009. Computer programs are used to receive signals from microscopes and combine thousands of two-dimensional images into an image 3. The exact dimension records the structure of the virus. According to Jiang, this requires the use of a large data system and work will not be possible without resources from the Purdue University IT office (ItaP).
Jiang used Purdue University's Condor program - a program that connects computers including desktop computers and research-specific computers - to create the largest computer network in any Which university.
He said: 'ItaP has provided us with the technological power to create the supercomputer system that is essential for this project. Purdue's Condor program allows us to take advantage of the power of 7000 computers. This is also a key factor in our success. ' Jiang intends to continue to improve the steps of the cycle to improve the ability of the technology to understand more viral strains in medicine.
Purdue University's biological group has studied a variety of issues, including: signaling pathways in cells, RNA catalysis, biological retrospective, viral entry pathways, viral copies and related diseases. to the virus. Researchers have combined X-ray crystallography, electron microscopy, NMR spectroscopy and other advanced computer-based mold making tools to understand the above problems.
The paper explaining the research is published in the March 28 issue of Nature .
- The computer increases the resolution for the image under the microscope
- NEC introduces 'ultra-resolution'
- The biggest virus in the world is exposed
- New drugs treat HIV and malaria
- Russia launches Earth image portal
- Photobie - Low-resolution PC image processing solution
- Check out the most dangerous viruses on the planet
- How do scientists capture a virus image?
- Take pictures from space capture the situation on the ground
- Knock off DNA to keep the viral gene inside
- The biggest star in the Milky Way is forming
- NASA released an image that recorded the Antares missile explosion last year
 Why do potatoes have eyes?
Why do potatoes have eyes? 'Tragedy' the world's largest carnivorous life: Death becomes ... public toilet
'Tragedy' the world's largest carnivorous life: Death becomes ... public toilet Tomatoes were once considered 'poisonous' for 200 years
Tomatoes were once considered 'poisonous' for 200 years Detecting microscopic parasites on human face
Detecting microscopic parasites on human face