Decode the smallest bacterial genome
1. The race to find the smallest biological genome has suddenly entered a new phase. The new work [1] published the ~ 422-kb genome of aphid bacterium, Buchnera aphidicola. However, Nakabachi et al [2] group published an even smaller genome of the bacterium bee symbiosis Carsonella ruddii. Thus, these are the two smallest bacterial genomes that have been decoded to date. On the other hand, these genomes are even equivalent to the geome size of chloroplasts (
2. Symbiotic relationships are relatively common ecological relationships among invertebrates, including important pests in agriculture and medicine. It is estimated that about 10% of the demand for nutrients such as cofactor, amino acid or other essential substances that insects cannot receive through food is provided by endosymbiotic bacteria in muscle. their body [3] .
The most concentrated target is B. aphidicola. This bacterium has produced all the non-substituted amino acids, except tryptophan, inside a group of specialized cells of the aphid body. The mealybugs of the mother had transmitted B. aphidicola to their descendants several hundred million years ago. In the evolution of this host-symbiont relationship, about 75% of B. aphidicola genomes have been removed to remain the current 600 - 700 kb genome [4], [5] . In this genome, only the minimum genes needed for bacterial growth within a closed ecosystem with the host remain. In fact, about 88% of this symbiotic enzyme can predict its function through network algorithms with biochemical reactions that simulate endosymbiotic physiological conditions.
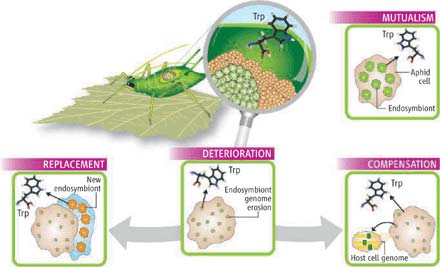
Evolutionary direction of endosymbiotic relationship (Photo: sciencemag)
3. Not only is the smallest bacterial genome, these two genomes are also the most stable genomes , since exogenous DNA is not required, does not contain repeating sequences exceeding 25 bp, and no recombinant recombination occurs. chromosomes range from 50 to 100 million years ago [6] . This suggests that this is an extremely sustainable biological system after gradually cutting down the symbiotic genome [7] . The rate of disappearance of a gene in the genome is estimated to take between 5 and 10 million years [8] . This is in line with Muller's conviction of gene loss mutations accumulated in small clones without the reception of new genes. As a result, such species can decline their vitality over time until completely extinct. It is now argued that the process of gene degeneration may stop or endosymbiotic genome will continue to scavenge causing the death of this species as well as the degradation of the their owners.
4. These two decoded geomes have surpassed the limits of the previous two decoding genomes of B. aphidicola, ranging in size from 615 to 641 kb. The new genome of B. aphidicola from the aphid Cinara cedri ( called B. aphidicola BCc strain) has a ~ 416 kb chromosome with 362 protein coding genes and a 6 kb round plasmid [9] . Meanwhile, the ~ 160 kb of C. ruddii chromosomes encode no more than 182 proteins (2). Both bacteria do not carry genes that encode most of the transport and transmembrane function proteins, this finding confirms that the free transport system follows passive diffusion pathways for metabolizing metabolites. ) between host and symbiotic. No gene was found to encode the tryptophan biosynthetic pathway on the BBc genome, although the experiment has shown that the aphid species depend on the source of tryptophan from bacteria [10] . The authors assume that the subsequent gene disappearance process in the endosymbiotic genome will be a lethal mutation for insects. Pérez-Brocal et al [11] proposed the possibility of escaping this situation by replacing the BBCc strain with a secondary symbiotic to provide tryptophan.
References:
V. Pérez-Brocal et al., Science 314, 312 (2006).
A. Nakabachi et al. Science 314, 267 (2006).
C. Dale, NA Moran, Cell 126, 453 (2006). [CrossRef]
S. Shigenobu et al., Nature 407, 81 (2000). [CrossRef]
I. Tamas et al. Science 296, [2376] (2002).
RCHJ van Ham et al., Proc. Natl. Acad. Sci. USA 100, 581 (2003). [CrossRef]
L. Klasson, SGE Andersson, Trends Microbiol 12, 37 (2004). [CrossRef]
C. Pal et al., Nature 440, 667 (2006). [CrossRef]
HJ Muller, Mutat. Res. 1, 2 (1964).
AE Douglas, WA Prosser, J. Insect Physiol. 38, 565 (1992). [CrossRef]
Koga, T. Tsuchida, T. Fukatsu, Proc. R. Soc. London B 270, 2543 (2003). [CrossRef]
ME Pettersson, OG Berg, Genetica, in press.
CG Kurland, SGE Andersson, Microbiol.Mol.Biol.Rev.64, 786 (2000).[Medline]
- Scientists 'fix' bacterial life trees
- Great turning point: People have 'programmed' bacteria
- Scientists successfully decode the spider's genome
- Successfully decode the genome of soybeans
- Decode the genome bamboo Moso
- Decode the 115-year-old woman's genome
- Decode the gorilla genome
- For the first time, Vietnam has successfully built the Vietnamese genome
- The first time to decode the DNA of kangaroo
- Successfully decode the Ebola virus genome
- Cooperation in human genome research
- The smallest smiley in the world
 Why do potatoes have eyes?
Why do potatoes have eyes? 'Tragedy' the world's largest carnivorous life: Death becomes ... public toilet
'Tragedy' the world's largest carnivorous life: Death becomes ... public toilet Tomatoes were once considered 'poisonous' for 200 years
Tomatoes were once considered 'poisonous' for 200 years Detecting microscopic parasites on human face
Detecting microscopic parasites on human face